Queens of the OceanJerome Hui snorkels in the genomic sea of jellyfish
September 2020

Jellyfish are not fish. They are the dancing queens of the ocean. If someone tells you the gelatinous creatures that move graciously in the ocean are in fact not too much different from the moths that fly and buzz in the air, your incredulity is perfectly normal. But if it comes from Prof. Jerome Hui, you better lend a patient ear.
Born, raised and educated in Hong Kong before he pursued a doctoral degree at Oxford University, Professor Hui, as most Hongkongers do, has first-hand knowledge of the Ocean Park which, incidentally, is the first theme park in Southeast Asia that opened its sea jelly exhibit. Little did he know that years later Ocean Park would turn a steady supplier of marine samples for his research after he had returned to Hong Kong and joined the School of Life Sciences of CUHK in late 2012.
Jellyfish belong in the phylum Cnidaria that includes sea anemones and corals. Those with a bell-shaped top and fluffy tentacles are in fact the adult (medusa) form of the species, often called true jellyfish. Its young (polyp) form is much smaller and resembles the sea anemones that are shelters to the popular clown fish.
Though a favourite among aquarium-goers, relatively little is known about jellyfish—how they evolve, how they reproduce, what they feed on, etc. One species, the Turritopsis dohrnii, is often nicknamed the immortal jellyfish which presents a biological puzzle as its medusa form under certain conditions would revert back to polyp, purportedly repeating the cycle in perpetuity, thus coming close to what is viewed as biological immortality.
Professor Hui has not examined an immortal jellyfish himself but thinks that it might have to do with the cell reprogramming in the jellyfish. But it is unmistakable that jellyfish undergo transformations in different developmental stages. Given his entomological interest, particularly in how insects morph (from egg to larvae to cocoon to insect), he is naturally drawn to the question of what enables jellyfish to go from, say, polyp to medusa. He is also curious about what makes jellyfish flourish in large numbers, so-called jellyfish blooms, which may pose dangers to humans and physical facilities. Swimmers and divers are well advised to stay away from the flotsam-like creatures, whose tentacles may release venom that stings them. A large number of jellyfish may also clog the cooling system of power facilities or wreak havoc on fishing apparatus.
Modern zoology is very much molecular zoology. The genomic make-up of the jellyfish is the first place to look for answers. This is what Professor Hui and his team have been doing for the past eight years. With uncommon perseverance and ingenuity they have broken new grounds in the understanding of this slippery creature. The seminal discoveries they made have recently been reported in an article published in Nature Communications. The article is so highly regarded that as at the time of writing it still appears as the featured article on the journal’s website.
Diving the genomic depth
Professor Hui and his team included two species of jellyfish in their study: the Amuska jellyfish (Sanderia malayensis) and one that sometimes found its way onto our plate—the flame jellyfish (Rhopilema esculentum). These species are commonly found in Asia waters.
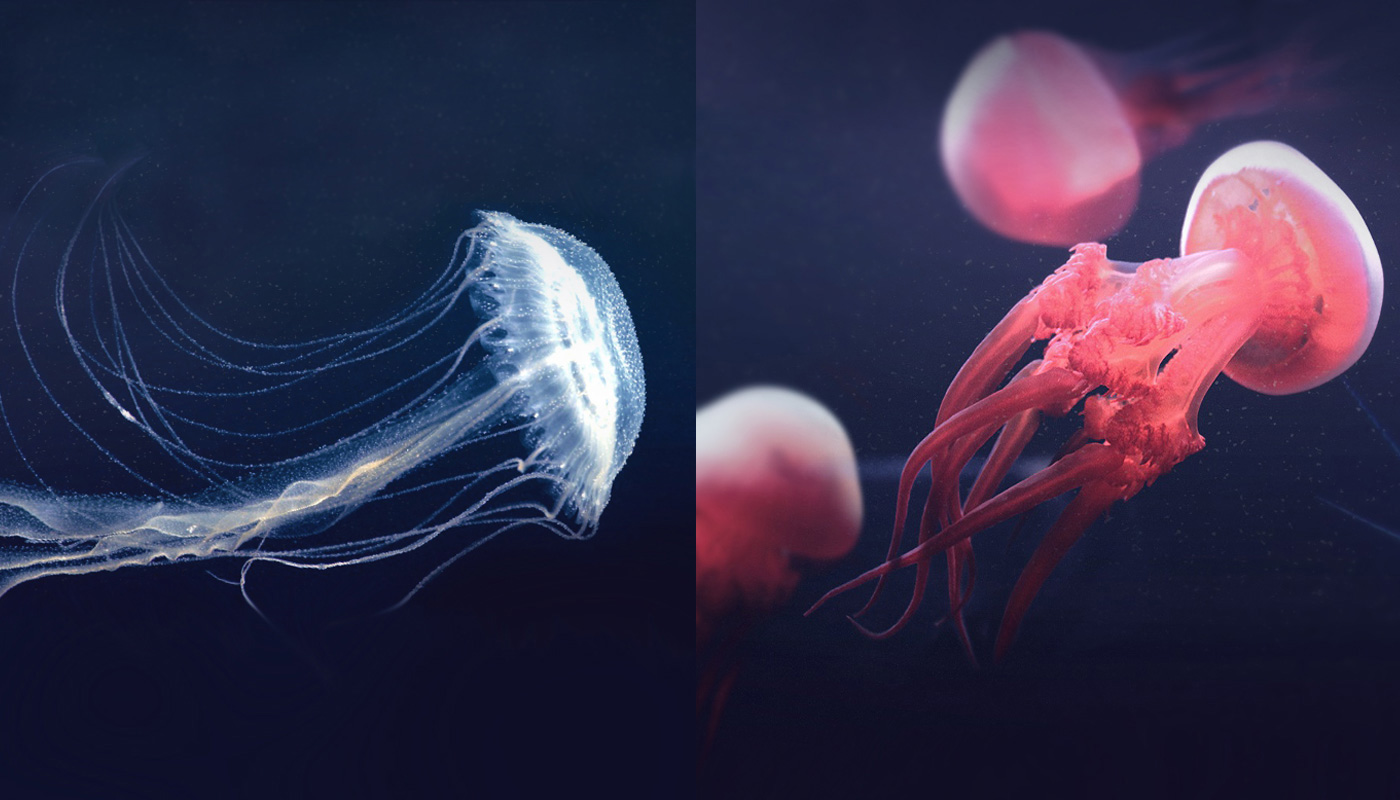
The first hurdle to overcome was to extract the DNA and RNA from the samples for genome sequencing. This turned out to be less straightforward than had previously been assumed, because over 90% of a jellyfish is made up of water. Conventional extraction techniques might fall short of the task. His team, however, went to great lengths and modified some protocols to finally get their hands on the DNA and RNA for sequencing and assembly.
In the course of genome sequencing and assembly, they had repeatedly innovated and improved on the techniques and protocols. The end results are two high-quality de novo reference genomes for the Amuska jellyfish and the flame jellyfish that reveal unique and conserved genomic features leading to insights into the evolutionary pathways of not only cnidarians but also bilaterians (animals with bilaterally symmetrical body structure), which make up 99% of extant animal species.
Change agent and common ancestry
In arthropods such as insects, transformations in form (the shedding of shell, the growth of wings, etc.) are regulated by the sesquiterpenoid (juvenile) hormones. As cnidarians including jellyfish also change forms during their lifecycles but no similar hormones had been found on them, it was thought that cnidarian metamorphosis must be governed by other unknown mechanisms. The high-quality de novo reference genomes assembled by Professor Hui and his team identify for the first time genes that encode the juvenile hormones in the Amuska and the flame jellyfish and hence settle the question. Future inquiry in the field can now proceed on the basis that genes for juvenile hormone production are present in cnidarians as well as in arthropods.
The ability to manipulate hormone production has many implications. Suppression of juvenile hormones in insects holds the key to the manufacturing of insecticides. It is not hard to see what practical applications the identification of similar juvenile hormones in jellyfish might have for, say, the viable control of jellyfish blooms.
The team also settled a dispute that had occupied scientists for over 20 years. The body of bilaterians including human beings can be differentiated into three germ layers—the ectoderm (skin and nervous system), the mesoderm (muscles) and the endoderm (guts), whereas that of cnidarians has only two—the ectoderm and the endoderm. The development of the three germ layers in bilaterians is regulated by what are known as the homeobox genes. What used to puzzle researchers was that homeobox genes are also found in cnidarians. In bilaterians, the homeobox genes have very conserved DNA sequences, i.e., the sequences have remained more or less the same across species throughout evolution. The homeobox genes in cnidarians, however, have divergent sequences, leading to the speculation that cnidarians did not inherit the homeobox genes from the same ancestor that bestowed the genes on the bilaterians but have evolved alone with genes only similar to those in the bilaterians. Professor Hui and his team found homeobox genes in the jellyfish which evidently belong in the bilaterians but have evolved differently. The implication is that both bilaterians and cnidarians share a common ancestor which, estimated to exist at least 600 million years ago, already had these important developmental genes.
The reference genomes of jellyfish assembled by Professor Hui and his team also shed light on two other issues concerning small RNAs and mitochondrial genomes. But these are holy grails for biologists rather than subjects for small talk among the uninitiated. As with all ground-breaking research, theirs gives rise to more questions than answers. Professor Hui sees great potential ahead and is confident that by leveraging on the strengths of CUHK in different departments and laboratories the way is paved for asking some hard questions of creatures that roam the land, soar in the air, or dance in the sea.
By tommycho@cuhkcontents


