CUHK
News Centre
CUHK Professor Lam Hon-Ming Announces World’s First Reference-grade Wild Soybean Genome
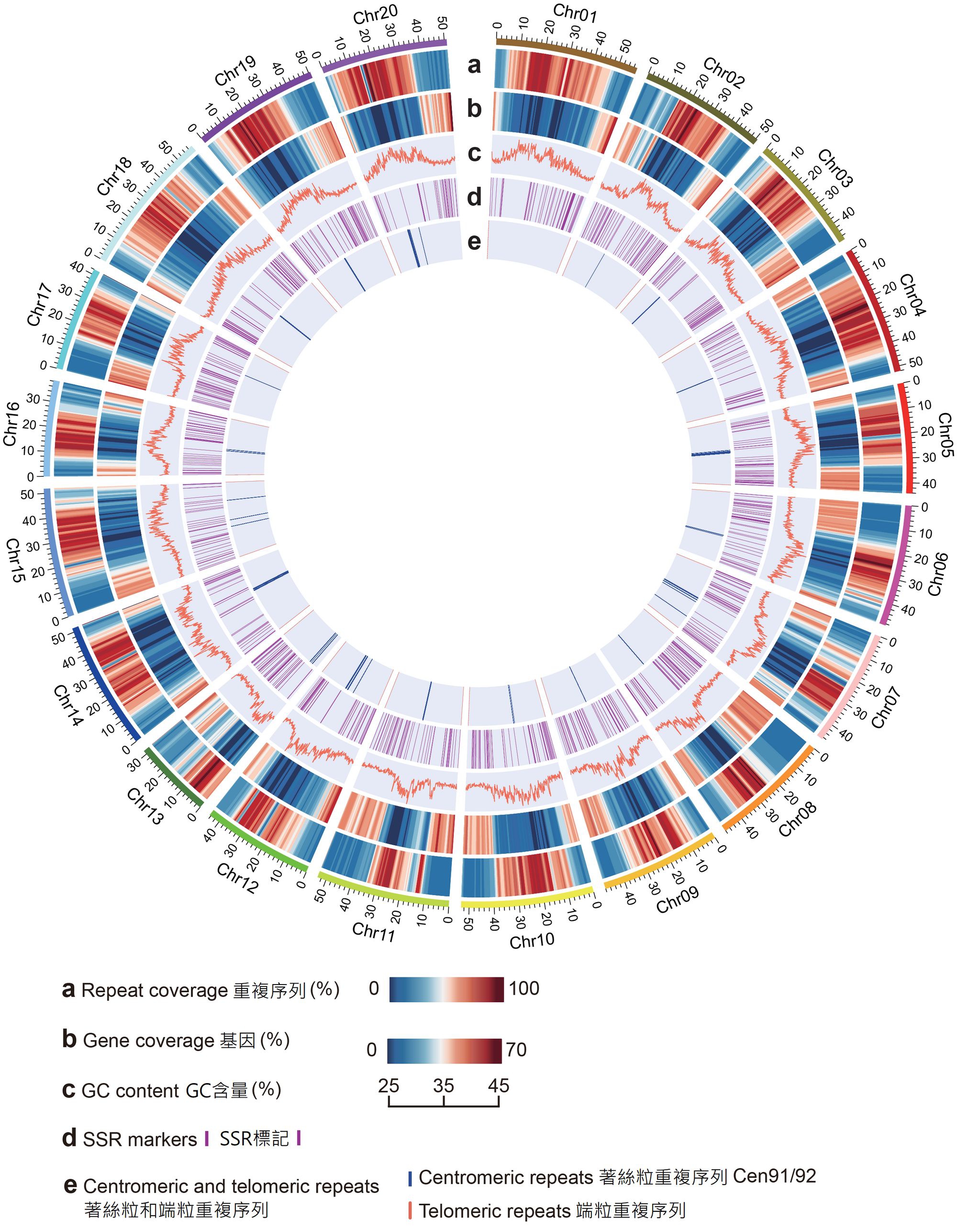
An international collaborative research team led by Professor Lam Hon-Ming, Director of the State Key Laboratory of Agrobiotechnology (The Chinese University of Hong Kong) and Professor of the School of Life Sciences at The Chinese University of Hong Kong (CUHK), completed the world’s first reference-grade wild soybean genome which provides an important tool for soybean genetic research internationally. It lays a solid foundation for comparative genomic studies of legume and soybean improvement programmes, and helps to improve the tolerance and other agronomic traits of soybean. It thereby extends the habitats of soybean cultivation and contributes to sustainable agriculture. The research findings have been published in the internationally renowned academic journal Nature Communications recently.
Professor Lam Hon-Ming said, “The reference-grade wild soybean genome has an important value in comparative genomics and evolutionary research. It can help to find important genes, and finally achieve improved varieties in cultivated soybean. This will help to develop high-yield, high-quality and high-tolerance soybean. I am sure that this reference genome will provide an important reference for other research teams dedicated to mining soybean genetic information.”
Soybean originated in China. After thousands of years of domestication, it has become an important cash crop and spread to other parts of the world. With the advances in science and the changing demand for agricultural technology, Professor Lam Hon-Ming took the lead in applying new genomics technology a decade ago. He found that wild soybean has richer biodiversity and genetic resources than cultivated soybean and that this can be used to improve agronomic traits of cultivated soybean, including stress tolerance, seed protein and secondary metabolite contents. However, in the past, genomic researches of soybean relied solely on the reference genome of the US-cultivated soybean Williams 82, which failed to effectively elucidate the genetic information that is specific to wild soybean.
Professor Lam’s research team adopted the third-generation sequencing technology provided by BGI Genomics to wild soybean W05, together with other new genome sequencing and bioinformatics techniques, to assemble the high-quality wild soybean reference genome at chromosome scale. Unlike the existing Williams 82 reference genome, researchers were able to identify significant structural differences between wild and cultivated soybean genomes. These differences have an important guiding significance in soybean gene mapping research and breeding practice. For example, a translocation between chromosome 11 and chromosome 13 was found. Such a translocation may affect the breeding between wild and cultivated soybeans, and have a strong impact on breeding strategies. At the same time, researchers discovered a large genomic inversion in the I locus area, which controls the seed coat colour. After rigorous experimental verification, it was confirmed that this inversion caused the switching from dark seeds to yellow seeds, which explained the long mystery in the process of soybean domestication.
Professor Lam Hon-Ming’s “Homecoming of Soybean” Project
Professor Lam Hon-Ming is a pioneer in soybean genomic and genetic research. He is committed to promoting sustainable agricultural development through his research, in line with the concept of climate-smart agriculture development advocated by the Food and Agriculture Organisation of the United Nations. In 2010, Professor Lam collaborated with BGI-Shenzhen to decode the genomes of 31 wild and cultivated soybeans, and concluded that wild soybean enjoys a much higher degree of genetic biodiversity. The finding was published as the cover story in Nature Genetics in December 2010, laying down the foundation for related research initiatives and projects, which have since been blooming in many areas of the world. This groundbreaking genomic research was conducted by an all-Chinese team in the “home” of soybean, which marks a disruption of the dominance of industrialised countries in high-end soybean research. The researchers have named the project “Homecoming of Soybean”.
Since then, Professor Lam’s team has been studying stress-tolerance genes in soybean with the use of state-of-the-art technology on genome sequencing and molecular biology. In 2014, Professor Lam successfully identified and cloned a novel salt-tolerant gene from wild soybean and applied it to the breeding of salt-tolerant soybean. The findings were published in Nature Communications in the same year. Using information from the salt-tolerance gene, Professor Lam worked with soybean breeders in the Mainland to generate soybean that can be grown on salinised arid land. Three new varieties of stress-tolerant soybean were successfully bred and passed the formal test in Gansu Province. The new varieties have been applied in semi-arid and drought-stricken areas in Northwest China. The team named the study “From Laboratory to Farmland”.
The newly published reference-grade wild soybean genome was completed by an international collaborative team led by Professor Lam, and the results will be applied to legume research and breeders all over the world. Thus the study was named “From China to the World”.
About the State Key Laboratory of Agrobiotechnology (The Chinese University of Hong Kong)
The State Key Laboratory of Agrobiotechnology (The Chinese University of Hong Kong) approved by the Ministry of Science and Technology of China is driven by basic research in agrobiotechnology, and adopts individual mature technology fields to applied basic research, with the goal of contributing to food security and climate smart agriculture in China and the world. The research focuses on: (1) the mining and application of important germplasm resources; (2) plant cell biology research; (3) drought tolerance and efficient agricultural water-saving research; (4) development of new technology platforms, including genomics, epigenetics, proteomics, animal development, and etc.