Rapid aneuploidy testing is widely implemented in routine prenatal diagnosis, it can be performed by molecular analysis of uncultured fetal cells. Quantitative fluorescence polymerase chain reaction (QF-PCR) analysis based on polymorphic small tandem repeat (STR) markers on chromosomes 13, 18, 21, X and Y has been successfully introduced, and its accuracy has been documented in a number of large-scale studies.
Test target: Aneuploidies of chromosomes 13, 18, 21, X and Y
Clinical information: Aneuploidies of chromosomes 13, 18, 21, X and Y are the most common chromosomal abnormalities detected in prenatal diagnosis, representing about 65% of all chromosomal abnormalities detected (Hassold et al. 1993).
Turn Around Time (TAT): 2 days
Specimen requirement:
| Sample type | Amount |
| Blood | 2-3 ml in EDTA bottles |
| Chorionic villi | 5 mg villi in transport medium |
| Amniotic fluid | 5-10 ml in sterile container |
| Placental tissues | At least 0.5ug minimal concentration of 30ng/uL; A260/A280 of ~1.8 |
Specimen submission:
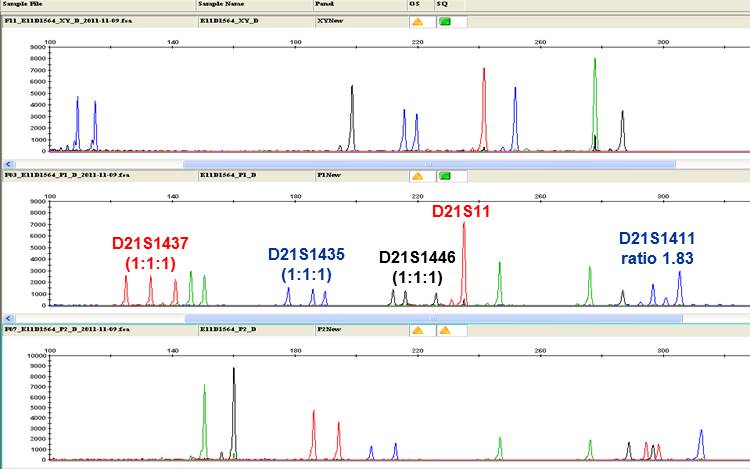
Fig. QF-PCR result of a carrying fetus of an individual consistent with Down syndrome (Trisomy 21). Four STR markers specific to chromosome 21 showed trisomic allele pattern.
Test target: Our in-house developed primer set covers most of the loci recommended by the best practice guidelines of the European Academy of Andrology (EAA) and the European Quality Monitoring Network Group (EMQN) 2004. The use of this primer set will enable the detection of over 95% of the deletions reported in the literature in the three AZF regions (Simoni et al. International Journal of Andrology, 2004; 27:240-249).
Clinical information: Y chromosomal microdeletions are the most common frequent genetic cause of male infertility. There are three recurrently deleted nonoverlapping subregions in the proximal, middle and distal Yq11 regions in the deleted interval designated as AZFa, AZFb and AZFc. Deletion of these loci results in spermatogenic arrest and is associated with azoospermia and oligozoospermia. Y chromosome infertility is characterized by azoospermia (absence of sperm), severe oligozoospermia (<1 x 106 sperm/mL semen), moderate oligozoospermia (1-5 x 106 sperm/mL semen), or mild oligozoospermia (5-20 x 106 sperm/mL semen). Males with Y chromosome infertility usually have no obvious symptoms, although physical examination may reveal small testes. According to our in-house data, PCR reveals microdeletions of the long arm of the Y chromosome in approximately 17% of these male.
Turn Around Time (TAT): 2 days
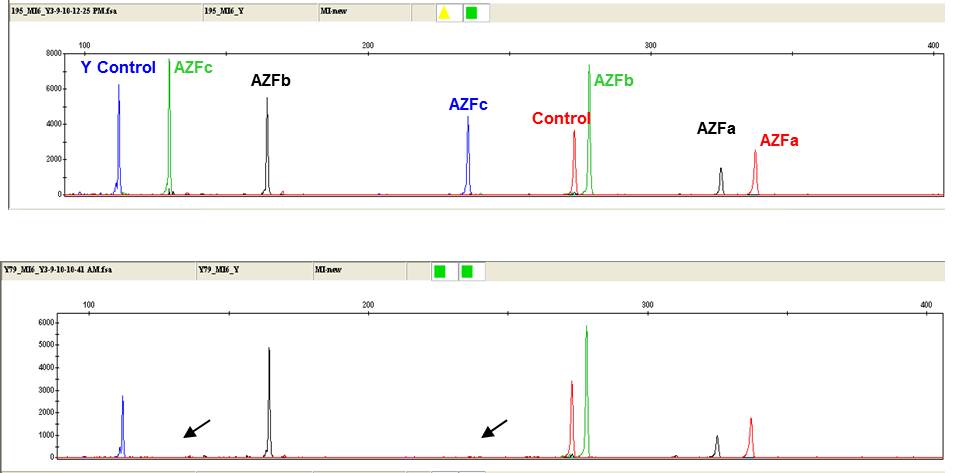
Fig. A representative electrophoretogram showing an individual consistent with AZFc region (lower panel) microdeletion.
Test target: polymerase chain reaction (PCR) using a pair of specific forward and reverse primers anneal to the upstream and downstream of the trinucleotide repeat region near the 5′ end of the FMR1 gene, followed by fragment analysis on a microfluidic capillary electrophoresis instrument and conversion of the measured PCR fragment size into the number of CGG repeats.
Clinical information: The diagnosis of Fragile X syndorme depends on the detection of an alteration in FMR1. More than 99% of individuals with fragile X syndrome have a loss-of-function mutation in FMR1 caused by an increased number of CGG trinucleotide repeats (typically >200) accompanied by aberrant methylation of FMR1. Other mutations within FMR1 that cause fragile X syndrome include deletions and point mutations. All individuals with Fragile X-associated tremor/ataxia syndrome (FXTAS) and FMR1-related primary ovarian insufficiency (POI) have FMR1 premutation trinucleotide repeats ranging from 55 to approximately 200.
Turn Around Time (TAT): 7 days
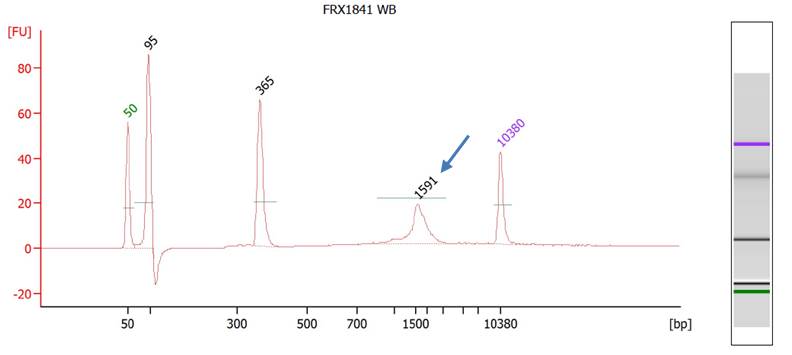
Fig. BioAnalyzer result of a female individual consistent with Fragile X full mutation carrier. Arrow indicates the expanded allele of FMR1.
Overview
Hearing impairment is one of the most common birth defects worldwide with the incidence of 1 in 500. In China, it has been estimated that over 27 million people are with hearing and speech disabilities. Of which more than 60% of these hearing impairment is caused by genetic factors. The most frequent mutations in non-syndromic hearing loss patients occur in GJB2, SLC26A4 and mitochondrial genome. Department of Obstetrics and Gynaecology, The Chinese University of Hong Kong offers hearing loss SNaPshot® genetic testing for screening of hearing impairment.
Test target: This test is a robust and efficient test that targets 14 common mutations on GJB2, SLC26A4 and mitochondrial DNA that are known to cause hearing loss, with carrier rate up to 1 in 6.3 babies.
Clinical information: Individuals in the following categories should consider Hearing Loss Screening:
- Universal Neonatal Hearing Screening positive
- Nonsyndromic hearing loss patients with unknown causes, excluding traumatic or infection reasons
- Normal individuals with family history of hearing impairment caused by these 14 mutations

Table. Carrier frequency of 14 common hearing loss mutations
Turn Around Time (TAT): 7 days
