FISH is a molecular cytogenetic technique developed in the early 1980s, aims to visualize and map the genetic material in an individual’s cells, including specific genes or portions of genes. Unlike conventional cytogenetic analysis, FISH does not require actively dividing cells, this make it a very versatile technique. Fluorescent probes are designed to bind to specific DNA sequence and the presence or absence of specific genetic regions can be visualized using fluorescence microscope.
Test target: This test detects numerical abnormalities of chromosomes 13, 18, 21, X and Y.
Clinical information: Aneuploidies of only 5 chromosomes (chromosomes 13, 18, 21, X and Y) account for about 65% of all chromosomal abnormalities and for 85-95% of the chromosomal aberrations causing live born birth defects.
Turn Around Time (TAT): 2-4 days
Specimen requirement:
| Sample type | Amount |
| Blood | 2-3 ml in heparin bottles |
| Chorionic villi | 5 mg villi in transport medium |
| Amniotic fluid | 5-10 ml in sterile container |
| Placental tissues | 5 mg tissues in transport medium |
Specimen submission:

Fig. CEP X & Y probes co-hybridized to interphase cells. Two green signals (Chr.X) and one red signal (Chr. Y) indicate Klinefelter syndrome (XXY)
Test target: This test detects microdeletion on chromosome 15q11.2 SNRPN gene locus (232Kb).
Clinical information: Approximately 70% of Prader-Willi/ Angelman syndrome patients demonstrate a deletion of 15q11.2-q13.
Turn Around Time (TAT): 2-4 days
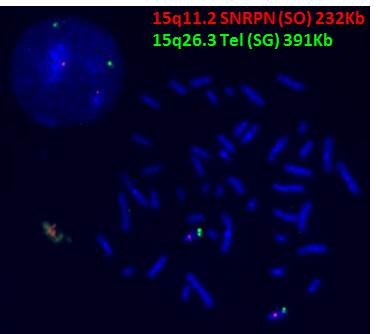
Fig. LSI SNRPN & Chr.15q Telomeric (control_) probes co-hybridized to metaphase and interphase cells. Two green signals (control_) and two red signals (SNPRN) indicate normal.
Test target: This test detects microdeletion on chromosome 22q11.2 TUPEL1 gene locus (232Kb).
Clinical information: Approximately 95% of 22q11.2 deletion syndrome is diagnosed in individuals with a submicroscopic deletion of chromosome 22 detected by fluorescence in situ hybridization (FISH), fewer than 5% of individuals with clinical findings of the 22q11.2 deletion syndrome have normal results on FISH testing due to individuals with atypical or nested deletions within the DGCR (DiGeorge chromosomal region) but not including the area encompassing the TUPLE FISH probe.
Turn Around Time (TAT): 2-7 days
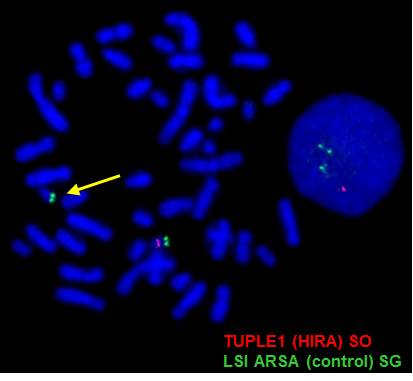
LSI TUPLE1 & ARSA (control_) probes co-hybridized to metaphase and interphase cells. Absence of the orange signal on one chromosome 22 (arrow) indicates deletion of the TUPLE1 locus at 22q11.2
Test target: For investigation of other targeted region please contact laboratory for probe availability.
