Single Cell Omics Core
About us
The SCOC provides a streamlined, cell-to-sequencing library service package for Single Cell RNA sequencing (scRNA-seq). It is capable to simultaneously profiling of transcriptomes ranging 500 – 10,000 cells in a single experiment. Different indexing systems are designed to label cell, transcript and sample origin in the sequencing library. This enables multiplexing of different sequencing libraries from multiple samples in the same Next Generation Sequencing (NGS) run. Libraries are Illumina-format compatible.
The service provided by SCOC includes an initial sample QC check, following by an evaluation of overall quality of sample for fitness to continue run. Single cells are then encapsulated in thousands of Gel Bead-in Emulsion (GEM) droplets, where cells are lysed and mRNA molecules reverse transcribed into cDNA. cDNA with specific cell barcodes are then pooled together and subjected to subsequent conventional sequencing library preparation. A final QC is performed on the final sequencing library to ensure sound quality for massively-parallel sequencing.
The SCOC welcomes interested PIs for experimental troubleshooting, intellectual advices as well as project discussions / collaborations.
Equipment

- Countess 3 Cell Counter (Cell viability check and counting)
- Light Microscope / Hemocytometer (Manuel cell counting)
- Compact cooling centrifuge
- 10X Controller (Single cell encapsulation)
- PCR workstation
- PCRs, thermomixer, vortex, plate centrifuge
- Tapestation 4200 (cDNA & sequencing library QC)
- Qubit 4 Fluorometer (DNA library quantification)
Service Workflow
Workflow overview

10X Chromium Next GEM Single Cell 3’ Gene Expression Workflow
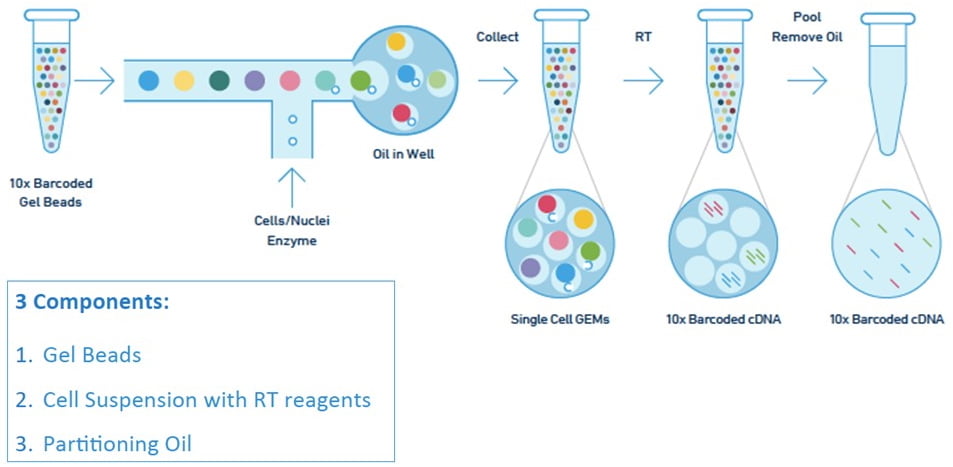
Chromium Next GEM Chip:

- Up to 8 samples per chip processed in parallel
- 500 to 10,000 cells per channel
- 18 minutes run time per chip
- Preferred cell size < 30 µm
- ~60-65 % cell recovery efficiency
RT, template switching

Pre-amp PCR

cDNA quality control
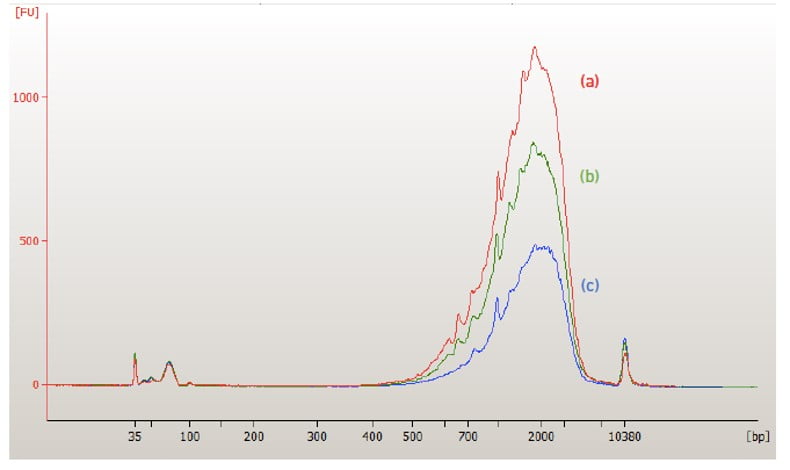
Sequencing library preparation

Sequencing library quality control

Service request
Please contact Dr Joaquim Vong (email: This email address is being protected from spambots. You need JavaScript enabled to view it. / 39430318) for service request, technical enquiry and project discussion.
For service request, please fill in our Sample submission form.
✓ Package set: From cell to sequencing library
✓ Cell quality and viability check / basic QC
✓ Sequencing library QC and quantification
✓ Intellectual advices (wet / dry lab)
✓ Troubleshooting
× Sequencing NOT included
× Repeated initial sample QC checks might incur extra charges
Output items
A. Sequencing library
B. QC reports for cDNA and sequencing library
C. Indexing information
The final product is a double-stranded cDNA molecule with sequencing adaptors and 3 different indices (10X barcode: cell info; UMI: transcript info; sample index: sample info). The sequencing library quality is evaluated by running into an Agilent 4200 TapeStation System.
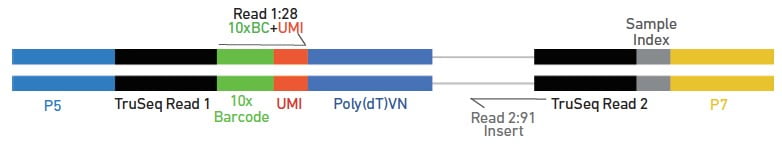
Source: 10X Genomics
Precautions
Users are required to fill in and sign the “Service Form for Single Cell Omics Core” prior to sample submission
https://webapp.sbs.cuhk.edu.hk/eform/view.php?id=55400
All users MUST declare the nature of sample(s) submitted to SCOC FREE of any bio-hazardous material.
An initial assessment of cell quality will be conducted for the generation of data with optimal quality. Users may take the risk of sub-optimal data quality if the initial cell quality check fails.
Upon service booking, a copy of the confirmed service form will be forwarded to both the end user as well as the corresponding PI.
Initial sample submission evaluation (sample quality, amount, etc) is recommended to optimize sample quality, depending on the sample nature. Potential experimental discussion(s) might be arranged prior to samples submission.
The complete workflow take approximately 2 full working days. As only limited amount of samples can be performed per week, users are encouraged to optimize their sample quality before submission. Service bookings are arranged on a first-come-first-serve basis.
Useful Resources
10X Genomics – Single Cell Gene Expression
https://www.10xgenomics.com/products/single-cell-gene-expression
10X University – A step-by-step learning and training environment
https://www.10xgenomics.com/10x-university
10X Datasets
https://www.10xgenomics.com/resources/datasets
10X Publications
https://www.10xgenomics.com/resources/publications
Worthington Tissue Dissociation Guide
http://www.worthington-biochem.com/tissuedissociation
Contacts
| Dr Joaquim Vong | |
| Scientific Officer | |
| Email: | This email address is being protected from spambots. You need JavaScript enabled to view it. |
| Tel no: | 39430318 |
| Miss Kathy Sham | |
| Technician | |
| Email: | This email address is being protected from spambots. You need JavaScript enabled to view it. |
| Tel no: | 39439859 |
| Location: | LIBSB Room 525 |
| Lab no.: | 39434373 |
