Unlocking DNA SecretsKevin Yip finds rogue genes in liver-cancer cells
May 2018

Every cell in our body contains the same DNA. But how cells behave is totally different. Why one cell becomes cancerous and another does not is a function of myriad interactions between genes and the enhancer ‘switches’ that turn them off or on.
It’s a complex web of relationships that Prof. Kevin Yip, an associate professor in CUHK’s Department of Computer Science and Engineering, is looking to explain.
In our genes, our DNA is responsible for the process that creates RNA and then proteins, the building blocks of the body. But it’s now clear that the non-coding parts of the DNA, much larger than the gene-transcription portion, regulate how the gene expresses itself.
Some of these segments act as enhancers, which activate a promoter ‘switch’ on target genes in the DNA, sometimes far away in the genome. The relationship is not always a direct one; in the makeup of mammals, there are far more enhancers than protein-coding genes. Enhancers often interact to produce different effects.
Professor Yip has used machine learning to incorporate a vast amount of data to infer which enhancers regulate which genes, and how they interact.
His work delved into an archive of 935 samples of human cells and tissue, some of which taken from the database FANTOM5 (Functional ANoTation of the Mammalian genome) of the Japanese research institute Riken. The samples come from many parts of the body, and are studied to map the human genome, which if depicted mathematically is a string of 3 billion characters.
There are some 25,000 genes in humans that code to create proteins. Even if we assume each gene has a binary ‘on’ or ‘off’ switch, the interactions of those genes would equate to 2 to the power of 25,000 possible combinations: a number larger than anything known in the universe.
Professor Yip’s study was the first to look at a broad array of enhancer-to-gene-target connections from different types of tissue. By studying both healthy and diseased tissue, the team homed in on evidence about how genes are regulated and why that mechanism sometimes goes wrong.
Scientists have often managed to identify sequences of DNA that are associated with specific diseases. But they do not know why, or if the sequence actually causes the disease, or is simply found at the same time.
Professor Yip ran a large amount of observations of cells in the 935 tissue samples through computer models to establish connections between the genes and gene enhancers in the DNA of different people and types of tissue. It requires a vast number of calculations to establish whether the various genes within one chromosome and their many enhancers are actually interacting with each other. To complicate matters, it is often necessary for more than one enhancer to be present for there to be any effect on a gene.
‘That’s just the first level: there could be false signal, the enhancer is there just by chance, or it may be controlling the gene indirectly,’ Professor Yip explains. ‘We try to make correlations, and build predictive models, so that we can tell up to a certain degree of accuracy whether the enhancers are really regulating the genes.’

Prof. Alfred Cheng, a liver-cancer expert in CUHK’s School of Biomedical Sciences, was able to guide Professor Yip in his work by identifying which genes are generally present in all cancers, vs. those that are specific to liver cancer.
Around 25 of the 935 samples were linked to the liver. Professor Yip began to compare ‘normal’ genes with the genes present in liver cancer, to see which enhancers were firing which promoters on which genes.
Professor Yip gradually narrowed his focus on liver-cancer cells, and succeeded in identifying three genes – PSRC1, RBM24 and TERT – that become hyperactive in that cancer due to a ‘perturbation’ or disturbance by different gene enhancers.
In many genes, a process called methylation suppresses the activity of the gene, preventing transcription from occurring and protein being formed.
In cancers, certain gene enhancers act abnormally and are de-methylated, causing the genes to activate. This can lead to the transcription of the gene, which may then create problem proteins. These ultimately can build into a tumor cell, and cancer. The calculations identified that the PSRC1, RBM24 and TERT genes are influenced in mutant liver cells by various enhancers and become hyperactive.
It’s possible to remove those enhancers, reverting the activation of the problematic genes. The team was able to edit the genome using an experimental method known as CRISPR/Cas9, which allows scientists to edit a DNA sequence by cutting out portions that include specific genes and enhancers to ‘see what happens.’ Therefore, they can test what happens when a specific enhancer is or is not present.
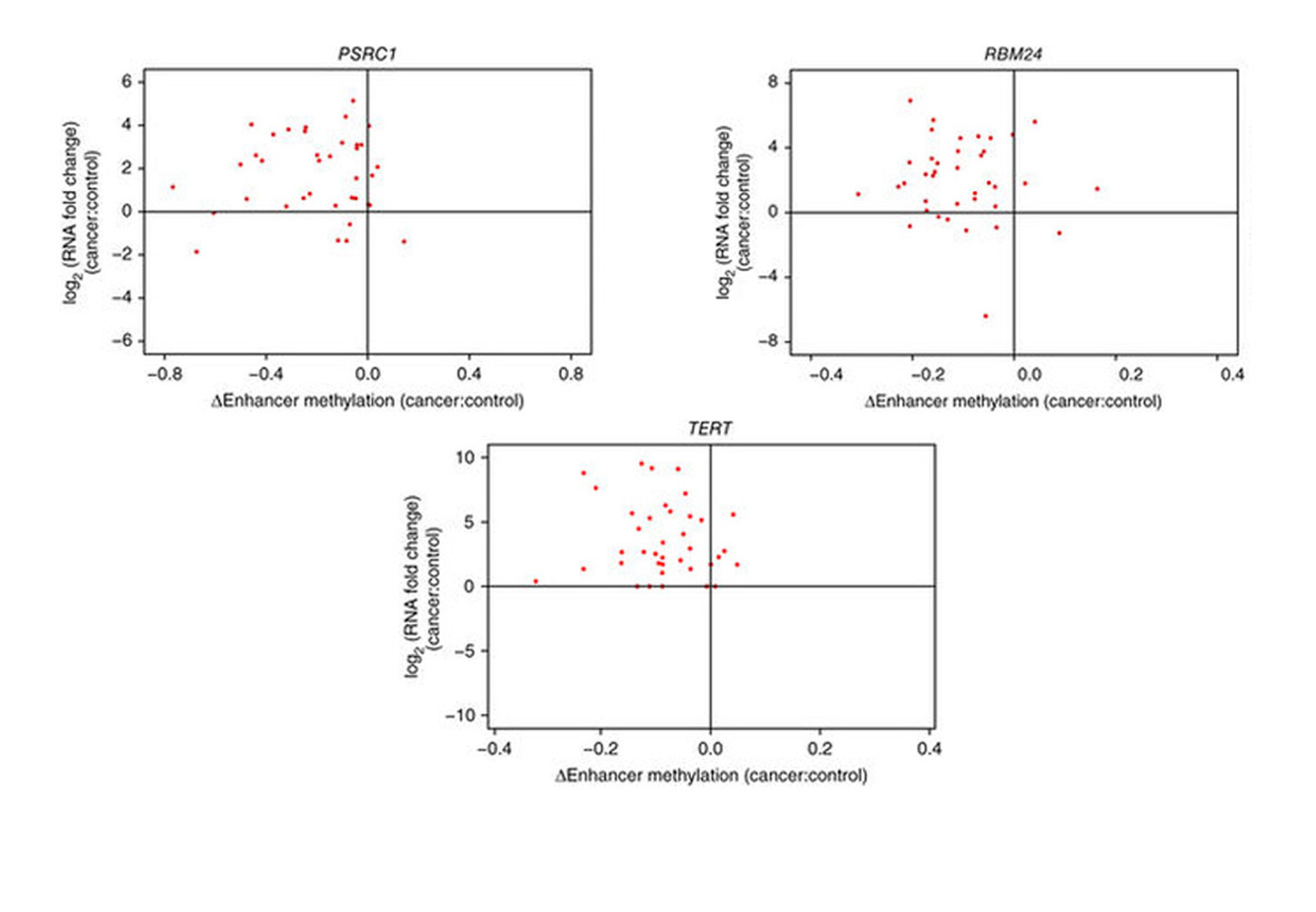
The team published the research last September in the journal Nature Genetics. Doctors could start to screen for various enhancers, to predict which patients are at risk of developing cancer. Scientists may then move on to create treatments such as drugs that disrupt the activity of the enhancer.
Processor Yip’s academic training is in computer engineering and computer science. In studying for his master’s degree, he began to work on biomedical research and bio-informatics, applying computational methods to solve medical problems.
In the laboratory, Professor Cheng’s team worked with data supplied by Professor Yip to test the hypotheses and validate the findings. Since even within one individual, some cells are cancerous and some are not, it requires Big Data computations to establish clear patterns of linkage and causation.
Computational testing takes time and effort. ‘In the long term, I hope I can cure cancer. But how long?’ Professor Yip jokes. ‘I hope I set the ball rolling so people can pick it up and really develop the relevant drug.’
By Alex Frew McMillan

