The Fruit Cracker
How Silin Zhong decodes fruition in nature

When a nutritionist bites an apple, its richness in vitamins, fibres, minerals, phytochemicals and antioxidants must be in her mind. When an evolutionary botanist bites an apple, she might take secret pride in her specialist knowledge that fleshy fruits only developed 80 million years ago and dry fruits have a much longer history. When Silin Zhong bites an apple, his enjoyment of the fruit is perhaps sweetened by his recent scientific finding of its ripening mechanism on the molecular level.
Fleshy fruits, though a latecomer in evolutionary terms, enjoy advantages in the propagation of the species over the dry varieties in that their nutrient-rich fleshy contents provide better incentives for animals to eat them and thereby spread the seeds to far-off places. Scientists have long wanted to fathom the mechanism which regulates the ripening of fleshy fruits. Like the gestation of a human foetus followed by labour, nature seems to have encoded when and under what conditions a fruit should ripen.
If the small prints of the genetic basis of this process can be made legible, we can understand better what happens and why despite the great variety in fruits they all seem to go through changes in size, texture, colour, taste and smell in ripening, followed by eventual decay.
Prof. Silin Zhong of CUHK’s School of Life Sciences leads an international team of scientists from the US, the UK, Spain and Argentina in the fruitENCODE project which aims to provide a comprehensive annotation of the functional elements in fleshy fruits in order to crack the mysterious process of fruit ripening.

The genomes of apple, pear, melon, peach, papaya, banana, watermelon, grape, strawberry, tomato and cucumber (the last two, though not often viewed as fruits, are indeed fruits in botanical terms) have been fully sequenced. Some of these fruits are climacteric fruits, so called because their ripening is facilitated by ethylene, a kind of plant hormone. Non-climacteric fruits do not depend on ethylene to ripen.
The fruitENCODE team studied the genetic and epigenetic bases of the evolution of these 11 fruits. By means of large-scale profiling of gene expression data and DNA methylation, and mapping of histone modifications and accessible chromatin regions, the team was able to identify three types of positive feedback loop that govern their fruit ripening processes. Their findings were recently published in Nature Plants.
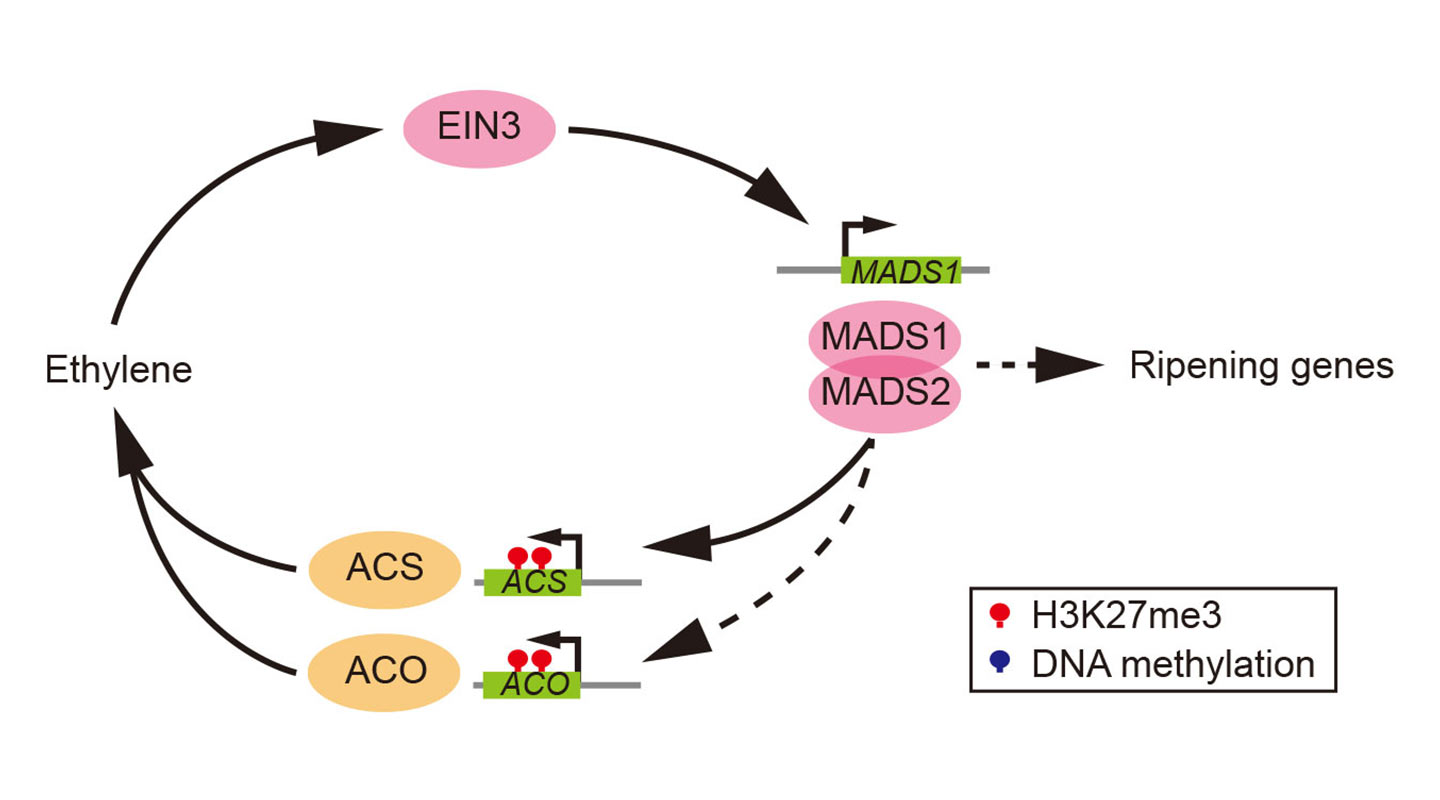
Apple, pear and tomato experienced recent whole genome duplication (doubling or even tripling their genome size) at the end of the Cretaceous period (145–66 million years ago). They evolved their fruits by using those duplicated floral organ identity genes (genes that specify the identity of the different organs of a flower) of their ancestors. In these fruits, a certain transcription factor (protein that controls if and when a gene gets activated) directly binds to genes that signal the production of ethylene and to others that relate to ripening characteristics such as softening and colour change. The result further triggers other transcription factors which come full circle to bind with the original transcription factor and so on, thereby starting a positive feedback loop that enables and sustains ethylene synthesis.
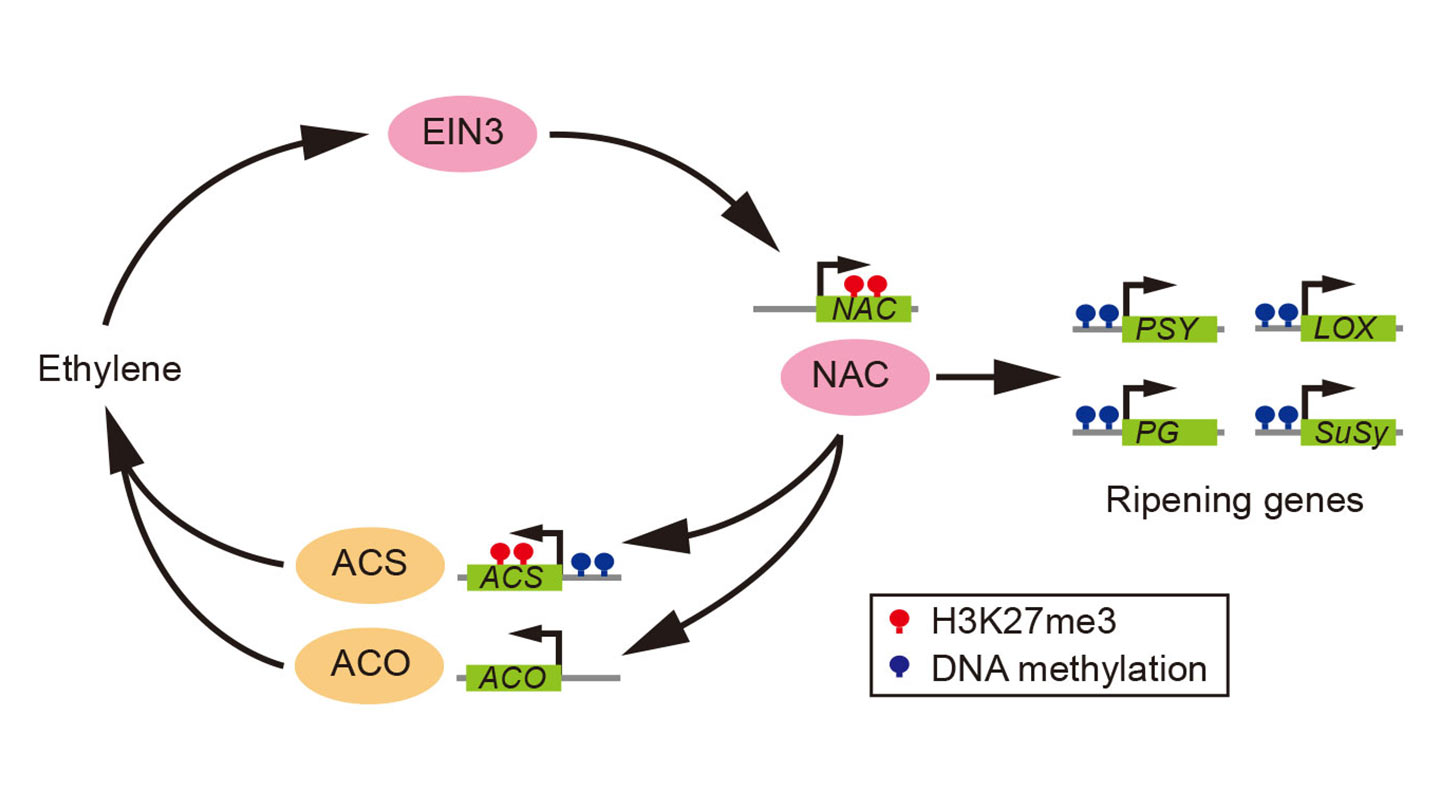
Melon, peach and papaya did not have the luxury of whole genome duplication during the late Cretaceous period, and therefore have not inherited the first group’s ethylene-production kit. They instead synthesize ethylene by converting their existing gene controlling senescence (ageing of the plant) to form a different positive feedback loop for the sustenance of ethylene synthesis.
The most eclectic of the bunch, banana, a climacteric monocot species with a history of genome duplications, reaches ripening by a combination of the two: using both the floral organ identity and the senescence genes. The non-climacteric fruits in the study—watermelon, cucumber, grape and strawberry—have developed their own system of ripening independent of ethylene.
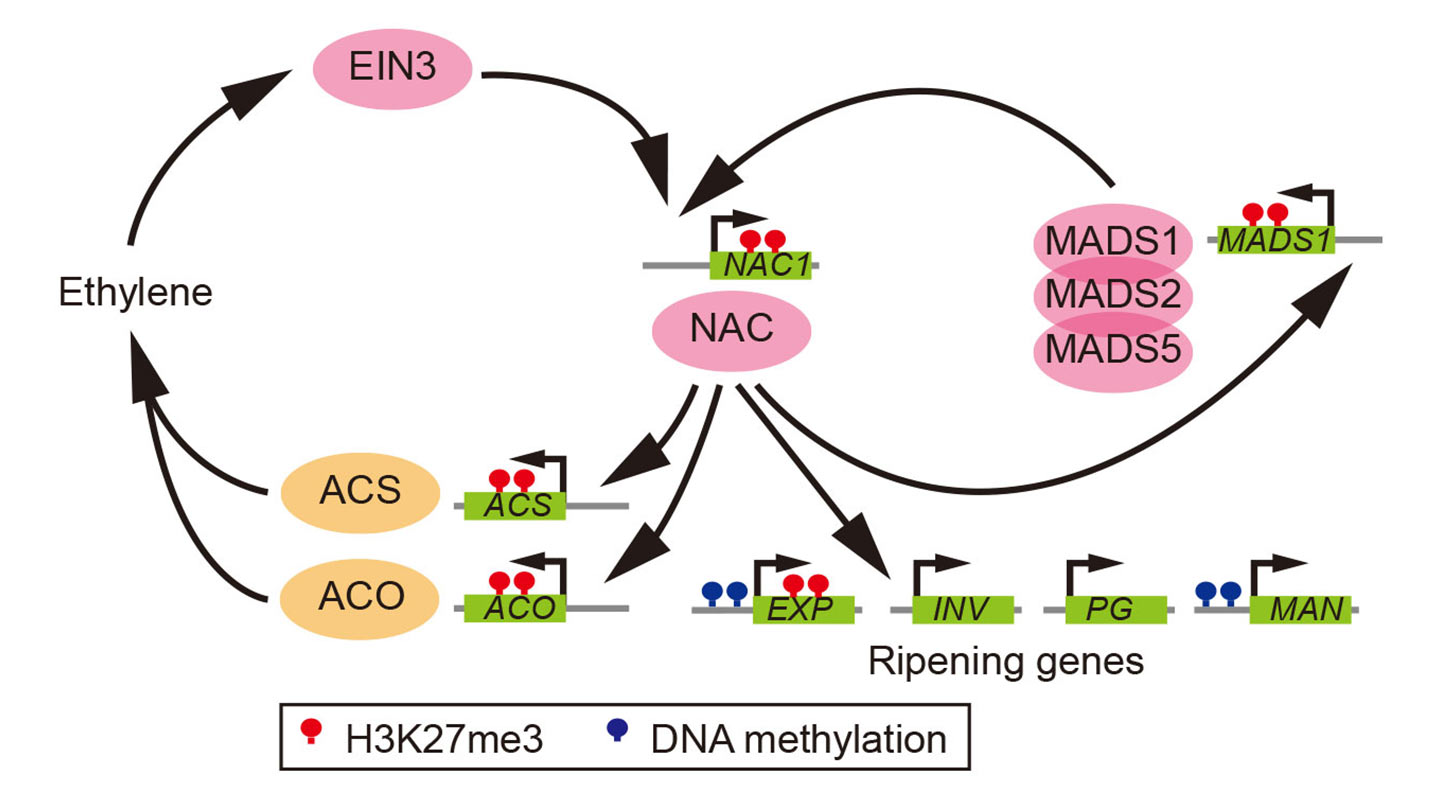
What the team also found is that the epigenetic mark H3K27me3, which represses key developmental genes in animals, plays an important regulatory role in the ripening process of fruits, too. It acts as a braking device that targets key ripening genes in plants to prevent premature ripening which may be undesirable in terms of the survival of the seeds.
Marks similar to H3K27me3 are found in the ripening genes of the ancestral plant species, suggesting that in their evolution, fruits like tomato, peach and banana have not just inherited the type of positive feedback loop from their ancestors but also their epigenetic marks to regulate ripening.
If the floral organ identity genes and the senescence genes are like data coded with information that bears upon ripening, H3K27me3 would be like software which tells when and how the genetic materials should behave. In Professor Zhong’s words: ‘Genome is like hardware; it is given and fixed. Epigenome, on the other hand, is like software; it can be written and rewritten.’
By elucidating the genetic and epigenetic mechanism of fruit ripening, the team’s findings could pave the way for a healthier and steadier food source in future. With greater understanding of the genomes of fruits and their ripening regulators, it is possible to manipulate and engineer the regulatory components in gene expressions to enhance their nutritional value, consumer appeal and shelf life. This is good news to fruit-lovers across the globe.
Professor Zhong loves fruits in general and cherry tomato and dragon fruit in particular. He may be looking to sink his teeth into their genomes next.
By T.C.
This article was originally published in No. 527, Newsletter in Nov 2018.

