CUHK and HKU Jointly Develop MARVEL Data Analysis Method to Detect Pathogenic Genes of Hirschsprung’s Disease
Hirschsprung’s disease is one of the most common congenital diseases requiring an abdominal operation in the newborn period. Its strong heritability suggests that genetic factors play a key role in the development of the disease. To find out genetic variants that may cause the disease, researchers from The Chinese University of Hong Kong (CUHK) and The University of Hong Kong (HKU) jointly developed a novel data analysis method called MARVEL (Multiscale Analysis of Regulatory Variants on the Epigenomic Landscape). A scientific article describing the MARVEL method and the corresponding findings about Hirschsprung’s disease has recently been published in the journal Genome Research.
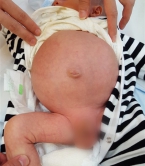
Most of us have experienced constipation before, which is caused by infrequent bowel movements. The situation can usually be improved within days by proper diet and physical exercise. A Hirschsprung patient has infrequent bowel movements starting from the day that he/she is born, and it can last his/her whole life if untreated, or even result in death. Typically characterised by a swollen belly, Hirschsprung patients need to go through a surgical operation to remove the “bad section” of the intestine and reconnect the healthy portions of the digestive system. Despite surgery, some patients continue to have infrequent bowel movements, or develop soiling and bowel infections. In Hong Kong, on average, there is a Hirschsprung case for every 3,500 newborns.
Scientists have long known that many Hirschsprung patients have mutations around a gene called RET. This gene codes for a protein that is important for a type of cells involved in gut development and function. Mutating the gene is analogous to damaging the mould, which causes the products produced (the proteins) to be malfunctioning. There are also mutations that do not change the protein but instead alter the amount of it produced in the cells.
Notwithstanding, only less than a quarter of the Hirschsprung patients have RET mutations. For the remaining majority, the cause of the disease is expected to be more complex and involves other lesser known genes. Since there are millions of genetic variants in every person, and most of them have nothing to do with the disease, it is difficult to pinpoint the variants and the corresponding affected genes that are functionally related to the disease. To find out these genes systematically, a joint research project by CUHK and HKU was established in 2014. It required substantial collaboration among different professions: medical doctors collected samples; genome scientists deciphered the DNA; geneticists characterised DNA differences between patients and non-patients; computer scientists developed mathematical models to identify the most critical DNA differences; biologists experimentally tested the significance of these computational findings.
MARVEL is one of the latest outcomes of the project. It is a computational data analysis method that has been used to discover many new genetic variants associated with Hirschsprung’s disease. Among its novelties, as compared to other methods, is its ability to consider the “convergent” effect of mutations; that the same functional impact can be produced by different mutations in different patients.
Professor Kevin Yip, Associate Professor of the Department of Computer Science and Engineering at CUHK, said, “This project would not have been possible without the collaborative effort of people from diverse backgrounds. It is becoming a basic requirement for high impact biomedical projects.”
Professor Yip is one of the three senior authors of the article in Genome Research. The other two are Professor Paul Tam and Professor Elly Ngan from the Department of Surgery at HKU.
Professor Tam explained that the novel computational method is an important first step towards the application of precision medicine to this heterogeneous congenital disorder. In future, genome data will likely become routine in the medical care of Hirschsprung patients, and computation based genome analysis can aid in stratifying patients to receive “tailor -made” and more effective treatments.
This project was funded by the Research Grants Council Theme-based Research Scheme (TRS). The project team will report its major research outcomes at the TRS Public Symposium on December 13th. Information about this symposium can be found at https://www.ugc.edu.hk/eng/rgc/about/events/symposium/symposium20.html.

The senior authors of the MARVEL project are Professor Paul Tam, Department of Surgery at HKU (middle, front row), Professor Elly Ngan, Department of Surgery at HKU (1st left, second row) and Professor Kevin Yip, Department of Computer Science and Engineering at CUHK (2nd left, third row).












 Subscribe
to Rss Feed
Subscribe
to Rss Feed Print
Print previous
previous



